- Cello CAD — Verilog-like compiler that emits DNA sequences. Github repo has more, and Science paper forthcoming.
- Privacy-Preserving Read Mapping Using Locality Sensitive Hashing and Secure Kmer Voting — crypographically preserved privacy when using cloud servers for read alignment as part of genome sequencing.
- How to Network in Five Easy Steps (Courtney Johnston) — aimed at arts audience, but just as relevant to early-career tech folks.
- Quantified Baby — The idea of self-tracking for children raises thorny questions of control and consent, Nafus said. Among hard-core practitioners, the idea has not really taken off, even as related products have started hitting the market.
"genomics" entries


Four short links: 4 April 2016
Verilog to DNA, Crypto Sequencing, How-To Network, and Quantified Baby


Four short links: 18 February 2016
Potteresque Project, Tumblr Teens, Hartificial Hand, and Denied by Data
- Homemade Weasley Clock (imgur) — construction photos of a clever Potter-inspired clock that shows where people are. (via Archie McPhee)
- Secret Lives of Tumblr Teens — teens perform joy on Instagram but confess sadness on Tumblr.
- Amazing Biomimetic Anthropomorphic Hand (Spectrum IEEE) — First, they laser scanned a human skeleton hand, and then 3D-printed artificial bones to match, which allowed them to duplicate the unfixed joint axes that we have […] The final parts to UW’s hand are the muscles, which are made up of an array of 10 Dynamixel servos, whose cable routing closely mimics the carpal tunnel of a human hand. Amazing detail!
- Life Insurance Can Gattaca You (FastCo) — “Unfortunately after carefully reviewing your application, we regret that we are unable to provide you with coverage because of your positive BRCA 1 gene,” the letter reads. In the U.S., about one in 400 women have a BRCA 1 or 2 gene, which is associated with increased risk of breast and ovarian cancer.


Four short links: 25 February 2015
Bricking Cars, Mapping Epigenome, Machine Learning from Encrypted Data, and Phone Privacy
- Remotely Bricking Cars (BoingBoing) — story from 2010 where an intruder illegally accessed Texas Auto Center’s Web-based remote vehicle immobilization system and one by one began turning off their customers’ cars throughout the city.
- Beginning to Map the Human Epigenome (MIT) — Kellis and his colleagues report 111 reference human epigenomes and study their regulatory circuitry, in a bid to understand their role in human traits and diseases. (The paper itself.)
- Machine Learning Classification over Encrypted Data (PDF) — It is worth mentioning that our work on privacy-preserving classification is complementary to work on differential privacy in the machine learning community. Our work aims to hide each user’s input data to the classification phase, whereas differential privacy seeks to construct classifiers/models from sensitive user training data that leak a bounded amount of information about each individual in the training data set. See also The Morning Paper’s unpacking of it.
- Privacy of Phone Audio (Reddit) — unconfirmed report from Redditor I started a new job today with Walk N’Talk Technologies. I get to listen to sound bites and rate how the text matches up with what is said in an audio clip and give feed back on what should be improved. At first, I though these sound bites were completely random. Then I began to notice a pattern. Soon, I realized that I was hearing peoples commands given to their mobile devices. Guys, I’m telling you, if you’ve said it to your phone, it’s been recorded…and there’s a damn good chance a 3rd party is going to hear it.


Four short links: 24 February 2015
Open Data, Packet Dumping, GPU Deep Learning, and Genetic Approval
- Wiki New Zealand — open data site, and check out the chart builder behind the scenes for importing the data. It’s magic.
- stenographer (Google) — open source packet dumper for capturing data during intrusions.
- Which GPU for Deep Learning? — a lot of numbers. Overall, I think memory size is overrated. You can nicely gain some speedups if you have very large memory, but these speedups are rather small. I would say that GPU clusters are nice to have, but that they cause more overhead than the accelerate progress; a single 12GB GPU will last you for 3-6 years; a 6GB GPU is plenty for now; a 4GB GPU is good but might be limiting on some problems; and a 3GB GPU will be fine for most research that looks into new architectures.
- 23andMe Wins FDA Approval for First Genetic Test — as they re-enter the market after FDA power play around approval (yes, I know: one company’s power play is another company’s flouting of safeguards designed to protect a vulnerable public).

Sequencing, cloud computing, and analytics meet around genetics and pharma
Bio-IT World shows what is possible and what is being accomplished
If your data consists of one million samples, but only 100 have the characteristics you’re looking for, and if each of the million samples contains 250,000 attributes, each of which is built of thousands of basic elements, you have a big data problem. This is kind of challenge faced by the 2,700 Bio-IT World attendees, who discover genetic interactions and create drugs for the rest of us.
Often they are looking for rare (orphan) diseases, or for cohorts who share a rare combination of genetic factors that require a unique treatment. The data sets get huge, particularly when the researchers start studying proteomics (the proteins active in the patients’ bodies).
So last week I took the subway downtown and crossed the two wind- and rain-whipped bridges that the city of Boston built to connect to the World Trade Center. I mingled for a day with attendees and exhibitors to find what data-related challenges they’re facing and what the latest solutions are. Here are some of the major themes I turned up.


Four short links: 25 February 2014
MtGox Go Boom, Flappy Bird, Air Hockey Hack, and Robo Lab
- Bitcoin Markets Down — value of bitcoins plunges as market uncertain after largest bitcoin exchange goes insolvent after losing over 750k bitcoins because they didn’t update their software after a flaw was discovered in the signing of transactions.
- Flappy Bird for the Commodore 64 — the 1980s games platform meets the 2014 game. cf the machine learning hack where the flappy bird learns to play the game successfully.
- Air Hockey Robot — awesome hack.
- Run 30 Lab Tests on Only One Drop of Blood — automated lab processing to remove the human error in centrifuging, timing, etc. that added to variability of results.

Big Data systems are making a difference in the fight against cancer
Open source, distributed computing tools speedup an important processing pipeline for genomics data
As open source, big data tools enter the early stages of maturation, data engineers and data scientists will have many opportunities to use them to “work on stuff that matters”. Along those lines, computational biology and medicine are areas where skilled data professionals are already beginning to make an impact. I recently came across a compelling open source project from UC Berkeley’s AMPLab: ADAM is a processing engine and set of formats for genomics data.
Second-generation sequencing machines produce more detailed and thus much larger files for analysis (250+ GB file for each person). Existing data formats and tools are optimized for single-server processing and do not easily scale out. ADAM uses distributed computing tools and techniques to speedup key stages of the variant processing pipeline (including sorting and deduping):
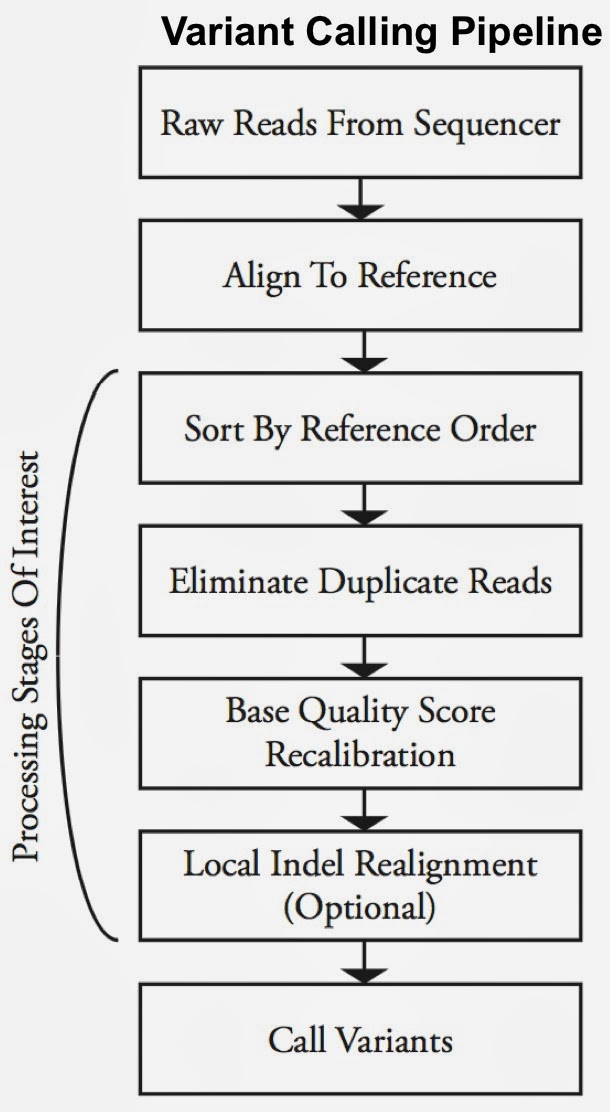
Very early on the designers of ADAM realized that a well-designed data schema (that specifies the representation of data when it is accessed) was key to having a system that could leverage existing big data tools. The ADAM format uses the Apache Avro data serialization system and comes with a human-readable schema that can be accessed using many programming languages (including C/C++/C#, Java/Scala, php, Python, Ruby). ADAM also includes a data format/access API implemented on top of Apache Avro and Parquet, and a data transformation API implemented on top of Apache Spark. Because it’s built with widely adopted tools, ADAM users can leverage components of the Hadoop (Impala, Hive, MapReduce) and BDAS (Shark, Spark, GraphX, MLbase) stacks for interactive and advanced analytics.

Ticking all the boxes for a health care upgrade at Strata Rx
What is needed for successful reform of the health care system?
Here’s what we all know: that a data-rich health care future is coming our way. And what it will look like, in large outlines. Health care reformers have learned that no single practice will improve the system. All of the following, which were discussed at O’Reilly’s recent Strata Rx conference, must fall in place.

Denny Ausiello discusses phenotypes, pathways, and stratification
A video interview with Colin Hill
Last month, Strata Rx Program Chair Colin Hill, of GNS Healthcare, sat down with Dr. Dennis Ausiello, Jackson Professor of Clinical Medicine at the Harvard Medical School, Co-Director at CATCH, Pfizer Board of Directors Member, and Former Chief of Medicine at the Massachusetts General Hospital (MGH), for a fireside chat at a private reception hosted by GNS. Their insightful conversation covered a range of topics that all touched on or intersected with the need to create smaller and more precise cohorts, as well as the need to focus on phenotypic data as much as we do on genotypic data.
The full video appears below.

Podcast: emerging technology and the coming disruption in design
Design's role in genomics and synthetic biology, robots taking our jobs, and scientists growing burgers in labs.
On a recent trip to our company offices in Cambridge, MA, I was fortunate enough to sit down with Jonathan Follett, a principal at Involution Studios and an O’Reilly author, and Mary Treseler, editorial strategist at O’Reilly. Follett currently is working with experts around the country to produce a book on designing for emerging technology. In this podcast, Follett, Treseler, and I discuss the magnitude of the coming disruption in the design space. Some tidbits covered in our discussion include:
- Design’s increasing role in genomics and synthetic biology. (For more on the genomic/synthetic biology space, here’s a recent Wired video interview with Craig Venter.)
- Robots taking our jobs, and what we humans will do for work.
- Embedded sensor networks and connected environments — soon, we’ll never get lost in a building again.
- Cross-pollination of industries to inform and evolve our emerging connected environments, such as the cross-disciplinary nature of the Wyss Institute for Biologically Inspired Engineering at Harvard.
- Approaching political policy as a design problem — politicians could benefit from design theory and rapid prototyping techniques found in design and manufacturing fields.
- Scientists growing burgers in labs.
And speaking of that lab burger, here’s Sergey Brin explaining why he bankrolled it:
Subscribe to the O’Reilly Radar Podcast through iTunes, SoundCloud, or directly through our podcast’s RSS feed.